6.1.1. Registration to common space¶
Here we show how to create a template from multiple anatomical scans and register all of them to it. Initially, registration is of extracted brains. Once these are reasonably aligned, whole heads are registered, weighted by masks that, if parameters are chosen well, include some scalp. The amount of scalp is hopefully enough to help in differentiating the brain-scalp boundary without including so much head tissue that it starts to contaminate the registration with the highly variable head tissue.
6.1.1.1. Retrieve the data¶
from sammba import data_fetchers
lemur = data_fetchers.fetch_lemur_mircen_2019_t2(subjects=[0, 1, 2])
retest contains paths to images and data description
print(lemur.anat)
Out:
['/home/salma/nilearn_data/mircen2019_t2/sub-01/sub-01_anat_sub-01_T2w.nii.gz', '/home/salma/nilearn_data/mircen2019_t2/sub-02/sub-02_anat_sub-02_T2w.nii.gz', '/home/salma/nilearn_data/mircen2019_t2/sub-03/sub-03_anat_sub-03_T2w.nii.gz']
6.1.1.2. Define the writing directory¶
import os
write_dir = os.path.join(os.getcwd(), 'lemur_common')
if not os.path.exists(write_dir):
os.makedirs(write_dir)
6.1.1.3. Create the template¶
from sammba.registration import anats_to_common
affine_register = anats_to_common(lemur.anat, write_dir, 400,
use_rats_tool=False, caching=True)
We set caching to True, so that the computations are not restarted. The paths to the registered images can be accessed easilly
registered_anats = affine_register.registered
print(registered_anats)
Out:
['/home/salma/CODE/sammba-mri/examples/01_template/lemur_common/sub-01_anat_sub-01_T2w_0_Unifized_for_brain_extraction_resample_shr_affine_catenated.nii.gz', '/home/salma/CODE/sammba-mri/examples/01_template/lemur_common/sub-02_anat_sub-02_T2w_1_Unifized_for_brain_extraction_resample_shr_affine_catenated.nii.gz', '/home/salma/CODE/sammba-mri/examples/01_template/lemur_common/sub-03_anat_sub-03_T2w_2_Unifized_for_brain_extraction_resample_shr_affine_catenated.nii.gz']
6.1.1.4. Assess the template¶
from nilearn import image
template_img = image.mean_img(registered_anats)
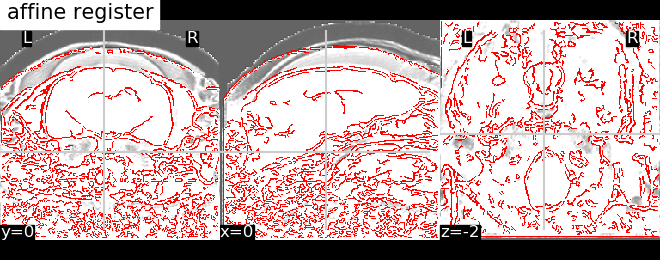
6.1.1.5. Visalize results¶
We plot the edges of one individual anat on top of the average image
from nilearn import plotting
average_img = image.mean_img(registered_anats)
display = plotting.plot_anat(average_img, dim=-1.6, title='affine register')
display.add_edges(registered_anats[0])
plotting.show()
6.1.1.6. Visualize pipeline steps¶
from sammba.graphs import create_pipeline_graph
graph_file = os.path.join(write_dir, 'affine_registration_graph')
create_pipeline_graph('anats_to_common_affine', graph_file)
Total running time of the script: ( 51 minutes 0.667 seconds)