6.2.1. Functional and anatomical coregistration¶
Standard functional preprocessing and registration of functional image to the anatomical.
6.2.1.1. Retrieve data¶
from sammba import data_fetchers
retest = data_fetchers.fetch_zurich_test_retest(subjects=[0],
correct_headers=True)
retest contains paths to images and data description
anat_filename = retest.anat[0]
func_filename = retest.func[0]
print(func_filename)
Out:
/home/salma/nilearn_data/zurich_retest/baseline/1366/rsfMRI_corrected.nii.gz
We use the Coregistrator, which coregisters the anatomical to a given modality
from sammba.registration import Coregistrator
coregistrator = Coregistrator(output_dir='animal_1366', brain_volume=400,
use_rats_tool=False, caching=True)
print(coregistrator)
Out:
Coregistrator(brain_volume=400, caching=True, clipping_fraction=0.2,
output_dir='animal_1366', use_rats_tool=False, verbose=True)
Coregistrator comes with a parameter clipping_fraction=.2 which sometimes needs to be changed to get a good brain mask. You can check how this parameter impacts the brain segmentation
from sammba.segmentation import brain_extraction_report
print(brain_extraction_report(anat_filename, brain_volume=400,
clipping_fractions=[.1, .2, .9, None],
use_rats_tool=False))
Out:
AP length RL width IS height symmetry volume
fraction 0.10 13.50 9.70 6.10 0.90 373.50
fraction 0.20 12.90 9.70 6.20 0.91 384.19
fraction 0.90 19.00 14.00 7.00 1.00 1862.00
no fraction 19.00 14.00 7.00 1.00 1862.00
6.2.1.2. Anatomical to functional registration¶
coregistrator.fit_anat(anat_filename)
coregistrator.fit_modality(func_filename, 'func', t_r=1.,
prior_rigid_body_registration=True)
The paths to the registered functional and anatomical images are accessible through the coregistrator attributes
registered_func_filename = coregistrator.undistorted_func_
registered_anat_filename = coregistrator.anat_in_func_space_
6.2.1.3. Check out the results¶
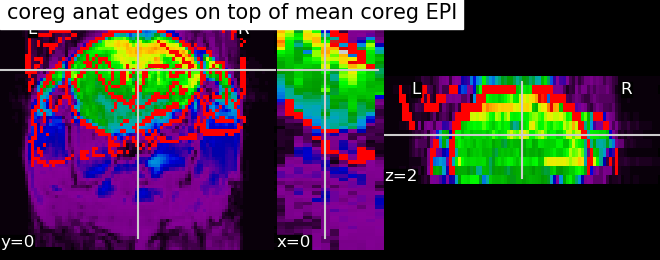
from nilearn import plotting, image
display = plotting.plot_epi(image.mean_img(registered_func_filename),
title='coreg anat edges on top of mean coreg EPI')
display.add_edges(registered_anat_filename)
plotting.show()
Total running time of the script: ( 0 minutes 50.045 seconds)