6.3.2. ICA on mouse¶
Independent components analysis on 5 mice.
6.3.2.1. Retrieve the fMRI data¶
from sammba import data_fetchers
retest = data_fetchers.fetch_zurich_test_retest(subjects=range(5),
correct_headers=True)
6.3.2.2. Load the template and its brain mask¶
dorr = data_fetchers.fetch_atlas_dorr_2008(downsample='100')
dorr_masks = data_fetchers.fetch_masks_dorr_2008(downsample='100')
print('Path to template is {} and to the brain mask is {}'.format(dorr.t2,
dorr_masks.brain))
Out:
Path to template is /home/salma/nilearn_data/dorr_2008/Dorr_2008_average_100um.nii.gz and to the brain mask is /home/salma/nilearn_data/dorr_2008/dorr_2008_brain_mask_100.nii.gz
6.3.2.3. Register to the template¶
import os
from sammba.registration import TemplateRegistrator
registrator = TemplateRegistrator(brain_volume=400, caching=True,
template=dorr.t2, use_rats_tool=False,
template_brain_mask=dorr_masks.brain,
registration_kind='affine')
registered_funcs = []
for anat, func in zip(retest.anat, retest.func):
animal_id = os.path.basename(os.path.dirname(anat))
registrator.output_dir = os.path.join('ica', animal_id)
registrator.fit_anat(anat)
registrator.fit_modality(func, 'func', t_r=1., voxel_size=(.3, .3, .3),
prior_rigid_body_registration=True)
registered_funcs.append(registrator.registered_func_)
6.3.2.4. Run ICA¶
Retrieve the independent components in brain space.
components_img = canica.masker_.inverse_transform(canica.components_)
6.3.2.5. Visualize the components¶
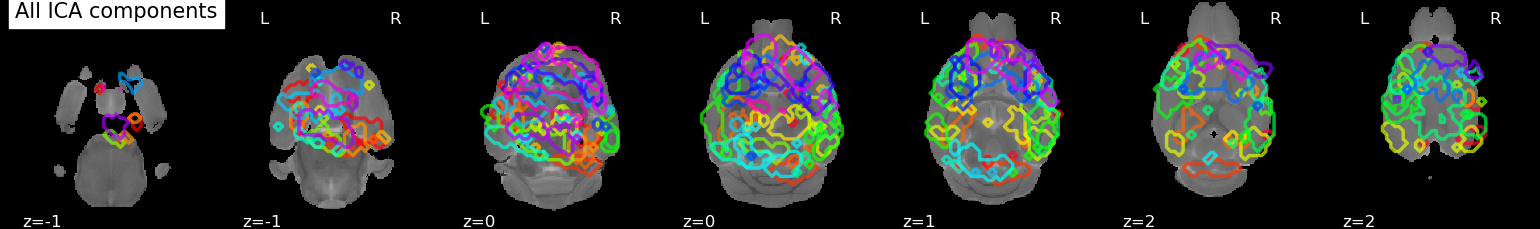
We can plot the outline of all components on one figure.
from nilearn import plotting
plotting.plot_prob_atlas(components_img,
bg_img=registrator.template_brain_,
display_mode='z',
title='All ICA components')
Total running time of the script: ( 8 minutes 31.678 seconds)